Introduction
This page is the continuation of my blog post on R commands. On the blog, see also why use R and the RSS feed of posts labelled R.
See also documentation at:
History of R
“Tukey defined data analysis in 1961 as:”Procedures for analyzing data, techniques for interpreting the results of such procedures, ways of planning the gathering of data to make its analysis easier, more precise or more accurate, and all the machinery and results of (mathematical) statistics which apply to analyzing data. Tukey’s championing of EDA encouraged the development of statistical computing packages, especially S at Bell Labs. The S programming language inspired the systems S-PLUS and R. This family of statistical-computing environments featured vastly improved dynamic visualization capabilities, which allowed statisticians to identify outliers, trends and patterns in data that merited further study.”
“S is one of several statistical computing languages that were designed at Bell Laboratories, and first took form between 1975–1976. Up to that time, much of the statistical computing was done by directly calling Fortran subroutines; however, S was designed to offer an alternate and more interactive approach, motivated in part by exploratory data analysis advocated by John Tukey. Early design decisions that hold even today include interactive graphics devices (printers and character terminals at the time), and providing easily accessible documentation for the functions.”
See also the comparison between R and Python below.
Control flow
Case when
dplyr::case_when is a generalised vectorised if. Example use:
x <- 1:50
case_when(
x %% 35 == 0 ~ "fizz buzz",
x %% 5 == 0 ~ "fizz",
x %% 7 == 0 ~ "buzz",
TRUE ~ as.character(x)
)For loop
https://cran.r-project.org/doc/manuals/r-release/R-intro.html#Repetitive-execution
for (x in c(“bla”, “bla”, “bli”)){ print(x) }
Data table
Database like operation benchmark between data table, dplyr, python pandas and other database like tools.
Data types
Data frames
The data type used in must analysis. Data frames are an aligned collection of vectors with observation in rows and variables in columns.
Size in memory
Print the size of a data frame in memory
print(object.size(df), units="MB")Vectors
Everything is a vector
There are no scalar values in R, only vectors, this makes it very natural to process times series or cross sectional data over many N observations as if we were processing only one instance of the variable.
SO answer concerning indexing up to end of vector/matrix “Sometimes it’s easier to tell R what you don’t want”
x <- c(5,5,4,3,2,1)
x[-(1:3)]
x[-c(1,3,6)]Set operations
x = letters[1:3]
y = letters[3:5]
union(x, y)## [1] "a" "b" "c" "d" "e"intersect(x, y)## [1] "c"setdiff(x, y)## [1] "a" "b"setdiff(y, x)## [1] "d" "e"setequal(x, y)## [1] FALSEDates
Create a date vector
as.Date("2022-01-01")
seq(as.Date("2022-01-01"), by=1, len=3)Parse a character vector into a date vector with lubridate
tralala_chr <- c("202012", "202101")
tralala <- lubridate::parse_date_time(tralala_chr, "%y%m")
tralalaRepresent the date object as a string
format(min(tralala),"%Y-%m")
format(max(tralala),"%Y-%m") Factors
Create factors
iris2 <- iris
iris2$species2 <- factor(iris2$Species, levels = c("setosa", "versicolor",
"virginica"), labels = c("A","B","C"))My answer to the question how to reorder factor levels the tidy way
If you happen to have a character vector to order, for example:
iris2 <- iris %>%
mutate(Species = as.character(Species)) %>%
group_by(Species) %>%
mutate(mean_sepal_width = mean(Sepal.Width)) %>%
ungroup()You can also order the factor level using the behavior of the forcats::as_factor function :
"Compared to base R, this function creates levels in the order in which they appear"
library(forcats)
iris2 %>%
# Change the order
arrange(mean_sepal_width) %>%
# Create factor levels in the order in which they appear
mutate(Species = forcats::as_factor(Species)) %>%
ggplot() +
aes(Species, Sepal.Width, color = Species) +
geom_point()Notice how the species names on the x axis are not ordered alphabetically but by increasing value of their mean_sepal_width. Remove the line containing as_factor to see the difference.
Strings
message("Using the following letters: ", paste(letters, collapse=","), ".")## Using the following letters: a,b,c,d,e,f,g,h,i,j,k,l,m,n,o,p,q,r,s,t,u,v,w,x,y,z.stringsAsFactors: An unauthorized biography
“The argument ‘stringsAsFactors’ is an argument to the ‘data.frame()’ function in R. It is a logical that indicates whether strings in a data frame should be treated as factor variables or as just plain strings. […] By default, ‘stringsAsFactors’ is set to TRUE.” “Most people I talk to today who use R are completely befuddled by the fact that ‘stringsAsFactors’ is set to TRUE by default. […]” “In the old days, when R was primarily being used by statisticians and statistical types, this setting strings to be factors made total sense. In most tabular data, if there were a column of the table that was non-numeric, it almost certainly encoded a categorical variable. Think sex (male/female), country (U.S./other), region (east/west), etc. In R, categorical variables are represented by ‘factor’ vectors and so character columns got converted factor. […] Why do we need factor variables to begin with? Because of modeling functions like ‘lm()’ and ‘glm()’. Modeling functions need to treat expand categorical variables into individual dummy variables, so that a categorical variable with 5 levels will be expanded into 4 different columns in your modeling matrix.” […]
“Around June of 2007, R introduced hashing of CHARSXP elements in the underlying C code thanks to Seth Falcon. What this meant was that effectively, character strings were hashed to an integer representation and stored in a global table in R. Anytime a given string was needed in R, it could be referenced by its underlying integer. This effectively put in place, globally, the factor encoding behavior of strings from before. Once this was implemented, there was little to be gained from an efficiency standpoint by encoding character variables as factor. Of course, you still needed to use ‘factors’ for the modeling functions.”
Levenshtein distance between words
Cf. https://en.wikipedia.org/wiki/Levenshtein_distance and
help(adist).
adist("kitten", "sitting")## [,1]
## [1,] 3Capitalise first letter
Capitalise the first letter of each word
stringr::str_to_title("bla bla bla")
# [1] "Bla Bla Bla"Lists
Given a list structure x, unlist simplifies it to produce a vector which contains all the atomic components which occur in x.
l1 <- list(a="a", b="2,", c="pi+2i")
str(l1)## List of 3
## $ a: chr "a"
## $ b: chr "2,"
## $ c: chr "pi+2i"unlist(l1) # a character vector ## a b c
## "a" "2," "pi+2i"str(unlist(l1))## Named chr [1:3] "a" "2," "pi+2i"
## - attr(*, "names")= chr [1:3] "a" "b" "c"Extract the first element of a nested list SO
minyearmaxyear <- list(c(2001, 2009), c(2010, 2014), c(2015, 2100))
library(purrr)
map(minyearmaxyear, 1)## [[1]]
## [1] 2001
##
## [[2]]
## [1] 2010
##
## [[3]]
## [1] 2015Remove items from a list
Remove elements from a named list in R.
l <- list(a = 1, b = 2)
l <- within(l, rm(a)) Remove elements from an unnamed list
l <- list(1,2)
l[1] <- NULLBind data frames in a list
# Large list if data frames
l_df <- list(head(iris), iris[c(92:95),], tail(iris))
df_stack <- data.frame()
for (i in 1:length(l_df)){
# Bind the first list item and remove it
df_stack <- rbind(df_stack, l_df[[1]])
l_df[1] <- NULL
}
# Reset row names
row.names(df_stack) <- NULLThis will take less memory than binding the data frame in this way:
l_df <- list(head(iris), iris[c(92:95),], tail(iris))
dfdt = data.table::rbindlist(l_df)And should give a similar result
identical(df_stack, data.frame(dfdt))
# TRUES3 methods
List all available methods for a class:
methods(class="lm") Debug
“NB: You shouldn’t need to use these tools when writing new functions. If you find yourself using them frequently with new code, reconsider your approach. Instead of trying to write one big function all at once, work interactively on small pieces. If you start small, you can quickly identify why something doesn’t work, and don’t need sophisticated debugging tools.”
Debug with base R
Print the call stack of the last error:
traceback()Start a browser at a particular position in code:
browser()Next, n: executes the next step in the function. If you have a variable named n, you’ll need print(n) to display its value.
Step into, or s: works like next, but if the next step is a function, it will step into that function so you can explore it interactively.
Finish, or f: finishes execution of the current loop or function.
Continue, c: leaves interactive debugging and continues regular execution of the function. This is useful if you’ve fixed the bad state and want to check that the function proceeds correctly.
Stop, Q: stops debugging, terminates the function, and returns to the global workspace. Use this once you’ve figured out where the problem is, and you’re ready to fix it and reload the code.
Debug with rlang
Print the call stack of the last error in simplified form:
rlang::last_error()Print the full call stack of the last error:
rlang::last_trace()More information on the behaviour of backtrace:
help(trace_back, package=rlang)“Because of lazy evaluation, the call stack in R is actually a tree, which the ‘summary()’ method of this object will reveal.”
Environnement
R environment
List objects in the global environment:
lsList objects coming from a specific loaded package:
ls("package:stats")
ls("package:stats", pattern="smooth")Remove all objects in the environment except one:
rm(list=ls()[!ls()=="object_to_keep"])
rm(list=ls()[!ls()=="con"]) # Remove all except a database connectionGet the current environment
rlang::current_env()Read system environment variables
Read an environment variable from the operating system
data_path <- Sys.getenv("PROJECT_DATA")Editors
I use Vim and the great Nvim-R plugin to edit R markdown documents and R scripts. I browse the table of content of R markdown files with the Voom plugin. I create R packages with Makefiles. I use the fugitive plugin to manage my git commits. See my Vim page for more information.
Rstudio is a great editors for those who prefer a graphical user interface. It has a Vim mode. In fact I trained my muscle memory for Vim movement keys inside RStudio initially. Rstudio also has a lot of menus and buttons to work with Rmarkdown documents, view the state of a git repository and build packages.
Input output
getwd()
list.files(tempdir())
dir.create("blabla")Apache Arrow and parquet files
“Read and write Feather files (read_feather(), write_feather()), a format optimized for speed and interoperability”
library(arrow)
read_parquet() # to read a single file
open_dataset() # to open a dataset sharded over multiple fileshelp(open_dataset)
” Call ‘open_dataset()’ to point to a directory of data files and return a ‘Dataset’, then use ‘dplyr’ methods to query it.”’
library(arrow)
library(dplyr)
# Set up directory for examples
tf <- tempfile()
dir.create(tf)
on.exit(unlink(tf))
data <- dplyr::group_by(iris, Species)
write_dataset(data, tf)
# open only the setosa part
setosa <- open_dataset(tf) %>%
filter(Species == "setosa") %>%
collect()CSV files
Read one csv file with default R function.
read.csv("data.csv", )Read many csv files with functions from the tidyverse packages.
First write sample csv files to a temporary directory. It’s more complicated than I thought it would be.
library(dplyr)
library(purrr)
library(purrrlyr)
library(readr)
data_folder <- file.path(tempdir(), "iris")
dir.create(data_folder)
iris %>%
# Keep the Species column in the output
# Create a new column that will be used as the grouping variable
mutate(species_group = Species) %>%
group_by(species_group) %>%
nest() %>%
by_row(~write.csv(.$data,
file = file.path(data_folder, paste0(.$species_group, ".csv")),
row.names = FALSE))Read these csv files into one data frame. Note the Species column has to be present in the csv files, otherwise we would loose that information.
iris_csv <- list.files(data_folder, full.names = TRUE) %>%
map_dfr(read_csv)write_csv returned an
Error in write_delim(...) is.data.frame(x) is not TRUE
That’s why we used write.csv instead.
RDS files
RDS is a Serialization Interface for Single Objects. Objects are
saved to binary data and compressed to the gzip format, see
help(saveRDS) for more details.
Write a dataset to an rds file:
saveRDS(iris, "/tmp/iris.rds")Read a dataset from an rds file
iris2 <- readRDS("/tmp/iris.rds")
identical(iris, iris2)Databases
Dplyr has databases backends to MySQl (MariaDB) and PostgreSQL.
Create directories
Create a directory do not show a warning if it exists already.
dir_path = "/tmp/blabla/"
dir.create(dir_path, showWarnings = FALSE)Find and replace
Longest contiguous stretch of non-NAs:
na.contiguous(c(NA,1:3,NA,NA,3:5,NA,NA))
] 1 2 3
tr(,"na.action")
] 1 5 6 7 8 9 10 11
tr(,"class")
] "omit"
tr(,"tsp")
] 2 4 1
na.contiguous(c(NA,1:3,NA,NA,3:6,NA,NA))
] 3 4 5 6
tr(,"na.action")
] 1 2 3 4 5 6 11 12
tr(,"class")
] "omit"
tr(,"tsp")
] 7 10 1Replace text in column names
Add the first row to the column name, useful for some double headed csv files coming from pandas.
paste(names(df), df[1,])Replace a regular expression with optional numerical pattern and final space
names(df) <- gsub(".per.hectare.[0-9]* *", "_", names(df))Using and, or in a regular expression
|Change columns to snake case
column_names <- c("Bla, Bla", "Hips, Hops", "Yield/Carcass Weight",
"Production", "Producing Animals/Slaughtered",
"Area harvested", "Yield")
gsub("[^[:alnum:]]","_",tolower(column_names))## [1] "bla__bla" "hips__hops"
## [3] "yield_carcass_weight" "production"
## [5] "producing_animals_slaughtered" "area_harvested"
## [7] "yield"# Replace one or more occurrences by an underscore
gsub("[^[:alnum:]]+","_",tolower(column_names))## [1] "bla_bla" "hips_hops"
## [3] "yield_carcass_weight" "production"
## [5] "producing_animals_slaughtered" "area_harvested"
## [7] "yield"iris2 <- iris
names(iris2) <- gsub("[^[:alnum:]]+","_",tolower(names(iris2)))
print(names(iris))## [1] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width" "Species"# [1] "sepal_length" "sepal_width" "petal_length" "petal_width" "species"Installing R and packages
To install R and Rstudio on Debian, see the debian.html#r_and_rstudio page on this site.
To install packages simply enter the following at an R command prompt:
install.packages("package_name")Some packages have dependencies that need to be installed at the OS level. Error messages:
"Configuration failed because libcurl was not found."
"Configuration failed because libxml-2.0 was not found."
"Configuration failed because openssl was not found."Can be solved by installing these dependencies:
sudo apt install libcurl4-openssl-dev
sudo apt install libxml2-dev
sudo apt install libssl-devInstall from source file
To install from a source file
install.packages("~/rp/FAOSTATpackage/FAOSTAT_2.2.1.tar.gz", repos=NULL)System information used by install.packages() to
determine default installation parameters
getOption("repos")
getOption("pkgType")Eurostat Package
Install required GDAL development library for the “sf” package
sudo apt install libgdal-devThen at the R prompt
install.packages("eurostat")Information about your R system
sessionInfo()
installed.packages()R Markdown
Create websites
Rmarkdown site generator
This website is generated from R Markdown documents with:
Rscript -e "rmarkdown::render_site()"The advantage of the rmarkdown site generator is that it creates pages with a floating, foldable table of content. It’s nice for pages that have a lot of subdivisions like this blog. The structure of a website is specified in a yaml file. For example, the code below creates a navigation bar:
navbar:
title: "Paul Rougieux"
left:
- text: "Home"
icon: fa-home
href: index.html
- text: "Tools"
icon: fa-wrench
menu:
- text: "Bash"
href: bash.htmlBookdown site generator
For blogs that fit into a book structure, with chapters, the bookdown package create a single clickable table of content for the whole site. It can also export the content to pdf format.
Specifying input from many sub directories in
_bookdown.yml:
rmd_files: ["index.Rmd", "chapters/chapt1.RMD", "chapters/chapt2.RMD"]Attention this will only work in the Merge and Knit strategy, as explained in the book chapter on two rendering approaches
“We call these two approaches “Merge and Knit” (M-K) and “Knit and Merge” (K-M), respectively.” “K-M does not allow Rmd files to be in subdirectories, but M-K does.”
Cross references
“Cross-referencing is not provided directly within the base rmarkdown package, but is provided as an extension in bookdown (Xie 2020c). We must therefore use an output format from bookdown (e.g., html_document2, pdf_document2, and word_document2, etc.) in the YAML output field.”
- Bookdown issue with flextable Table and figure cross references in word Now solved with the following implementation
iris %>% head() %>% flextable()This is a reference to flextable @ref(tab:irishead). The chunk name should not contain an underscore, otherwise the cross reference will not work. See bookdown issue 941.
See more flextable caption examples in
system.file(package = "flextable", "examples", "rmd", "captions")
Flextable and officer package
The officer package makes it possible to create improved word and powerpoint documents. The flextable package has a few functions to generate tables for word documents (otherwise they look funny).
Before installing flextable, the following system dependencies were required:
sudo apt install libcairo2-dev libjpeg-dev libgif-devInstallation in R:
install.packages("flextable")This is to work with colleagues who are not in the latex/pdf world.
Landscape pages
library(officer)
library(flextable)
ft_iris <- iris %>%
head() %>%
flextable() %>%
autofit()
# Add a flex table in landscape format
# Supply document name here, otherwise start an empty doc
doc <- read_docx() %>%
# Portait and landscape sections are defined by their ending
# A portrait section is ending here
body_end_section_portrait() %>%
body_add_flextable(value = ft_iris, split = TRUE) %>%
# A landscape section is ending here
body_end_section_landscape() %>%
print(target = "/tmp/iris.docx")kable to create tables
A table caption
knitr::kable(head(iris), caption = "An example table caption.")| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| 5.4 | 3.9 | 1.7 | 0.4 | setosa |
Examples copie from yihui’s R markdown cookbook.
set.seed(100)
d <- cbind(X1 = runif(3),
X2 = 10^c(3, 5, 7),
X3 = rnorm(3, 0, 1000))
# at most 4 decimal places
knitr::kable(d, digits = 4)| X1 | X2 | X3 |
|---|---|---|
| 0.3078 | 1e+03 | -1585.8810 |
| 0.2577 | 1e+05 | -40.6920 |
| 0.5523 | 1e+07 | -331.0045 |
# round columns separately
knitr::kable(d, digits = c(5, 0, 2))| X1 | X2 | X3 |
|---|---|---|
| 0.30777 | 1e+03 | -1585.88 |
| 0.25767 | 1e+05 | -40.69 |
| 0.55232 | 1e+07 | -331.00 |
# do not use the scientific notation
knitr::kable(d, digits = 3, format.args = list(scientific = FALSE))| X1 | X2 | X3 |
|---|---|---|
| 0.308 | 1000 | -1585.881 |
| 0.258 | 100000 | -40.692 |
| 0.552 | 10000000 | -331.005 |
# add commas to big numbers
knitr::kable(d, digits = 3, format.args = list(big.mark = ",",
scientific = FALSE))| X1 | X2 | X3 |
|---|---|---|
| 0.308 | 1,000 | -1,585.881 |
| 0.258 | 100,000 | -40.692 |
| 0.552 | 10,000,000 | -331.005 |
The format is recognised automatically by knitr based on the nature of the output document.
cat(kable(head(iris, 1), format = "html"))## <table>
## <thead>
## <tr>
## <th style="text-align:right;"> Sepal.Length </th>
## <th style="text-align:right;"> Sepal.Width </th>
## <th style="text-align:right;"> Petal.Length </th>
## <th style="text-align:right;"> Petal.Width </th>
## <th style="text-align:left;"> Species </th>
## </tr>
## </thead>
## <tbody>
## <tr>
## <td style="text-align:right;"> 5.1 </td>
## <td style="text-align:right;"> 3.5 </td>
## <td style="text-align:right;"> 1.4 </td>
## <td style="text-align:right;"> 0.2 </td>
## <td style="text-align:left;"> setosa </td>
## </tr>
## </tbody>
## </table>cat(kable(head(iris, 1), format = "latex"))##
## \begin{tabular}{r|r|r|r|l}
## \hline
## Sepal.Length & Sepal.Width & Petal.Length & Petal.Width & Species\\
## \hline
## 5.1 & 3.5 & 1.4 & 0.2 & setosa\\
## \hline
## \end{tabular}cat(kable(head(iris, 1), format = "markdown"))## | Sepal.Length| Sepal.Width| Petal.Length| Petal.Width|Species | |------------:|-----------:|------------:|-----------:|:-------| | 5.1| 3.5| 1.4| 0.2|setosa |Booktabs
Booktabs tables look nicer in latex document, they can be created by
specifying the argument booktabs = TRUE into the
kable() call.
Knitr adds an extra space every five lines into booktabs tables for
readability. This is undesirable for short tables. To remove this space
specify the linesep = "" argument to kable()
as explained in this SO
answer.
KableExtra formatting
The package kableExtra can group
rows via multi-row cells
Html format
library(dplyr)
iris3 <- iris %>% group_by(Species) %>% slice(1:3)
iris3 %>%
knitr::kable(format = "html") %>%
kableExtra::collapse_rows(columns = 5, valign = "top")Latex, pdf format
iris3 %>%
knitr::kable(format = "latex") %>%
kableExtra::collapse_rows(columns = 5, valign = "top")Make wider columns
iris %>%
knitr::kable(format = 'markdown', caption = 'Flower measurements') %>%
kableExtra::column_spec(5, width = "30em")
`Output
The default output is in HTML. The nature of the notebook output can be defined in the preamble section of the R markdown document.
It’s often useful to have a table of content at the start of long documents.
---
title: "Title"
date: "01 August, 2025"
output:
pdf_document:
number_sections: yes
toc: yes
---I usually have this line at the beginning of R markdown files to generate the pdf output from command line:
cd ~/rp/path_to_repo/notebooks/ && Rscript -e "rmarkdown::render('file_name.Rmd', 'pdf_document')"Power point documents
It’s possible to specify a power point template in the knitr preamble as such:
---
title: "Title"
output:
powerpoint_presentation:
reference_doc: presentation_template.pptx
---More details in https://bookdown.org/yihui/rmarkdown/powerpoint-presentation.html I ran into some issues: - https://stackoverflow.com/questions/56243502/how-to-fix-could-not-find-shape-for-powerpoint-content-error-pandoc-document-c - https://github.com/jgm/pandoc/issues/5402 And the fact that I cannot edit the slide master from power point online is annoying.
Figure numbering
Using bookdown it’s possible to use figure numbering as explain in this SO answer
---
output: bookdown::html_document2
---
# Section name {#id}
<div class="figure">
<img src="R_files/figure-html/pressure-1.png" alt="test plot" width="672" />
<p class="caption">test plot</p>
</div>
To cross-reference the figure, use `\@ref(fig:pressure)`
to produce Figure \@ref(fig:pressure). All this is found within the section \@ref(id).This doesn’t work with word documents because the word document format is not available for bookdown.
Word document
Render an Rmarkdown notebook as word document with the rmarkdown package:
Rscript -e "rmarkdown::render('chapter.Rmd', output_format='word_document')"Sample Rmarkdown document to be rendered with quarto
---
title: "My Document"
format:
docx:
toc: true
---
# Section name
<div class="figure">
<img src="R_files/figure-html/fig-plot-1.png" alt="Plot" width="672" />
<p class="caption">Plot</p>
</div>
For example, see @fig-plot.Render with:
quarto render document.qmdCreate a template:
quarto pandoc -o custom-reference-doc.docx \
--print-default-data-file reference.docxOther notes:
- The redoc package was developped in 2019, but abandoned. https://bookdown.org/yihui/rmarkdown-cookbook/word-redoc.html
Python engine
Rstudio: R Mardown python engine
See also the reticulate package mentioned below.
Reticulate to call python from R
To run a python script that prepares data
py_run_file("script.py")The object created by the python script will then be available from R
through the py$oject_name interface.
To run an import statement in python, then use the imported object from R.
library(reticulate)
py_run_string("from biotrade.faostat import faostat")
wheat <- py$faostat$db$select("crop_production", product="wheat")More information in the vignette:
vignette("calling_python", package="reticulate")Reticulate in a notebook
Example of a python chunk followed by an R chunk.
# ---
# title: "The Ultimate Question"
# ---
# ```{r setup}
# library(reticulate)
# ```
# ```{python}
# import pandas
# df = pandas.DataFrame({'x':[2,3,7], 'y':['life','universe','everything']})
# ```
# ```{r}
# str(py$df)
# prod(py$df$x)
# ```This can also be used to call python functions in R
λ{python eval=FALSE}
from biotrade.faostat import faostat
—
``` r
soy_prod <- py$faostat$db$select(table="crop_production", product = "soy")
```Using Reticulate in an R packages
“When you do this, you should use the delay_load flag to the import() function, for example:”
# global reference to scipy (will be initialized in .onLoad)
scipy <- NULL
.onLoad <- function(libname, pkgname) {
# use superassignment to update global reference to scipy
scipy <<- reticulate::import("scipy", delay_load = TRUE)
}“Using the delay_load flag has two important benefits:”
“It allows you to successfully load your package even when Python / Python packages are not installed on the target system (this is particularly important when testing on CRAN build machines).”
“It allows users to specify a desired location for Python before interacting with your package.”
Example R packages that use reticulate
Reproducibility and cloud computing
Cloudyr project
The cloudyr project is a collection of R packages to enable cloud computing.
R packages that are actively maintained can be seen on the github project page of cloudyr.
For example the googleCloudVisionR package gets the following image annotations for a picture of golden retriever puppies
description score topicality Dog 0.9953705 0.9953705 Mammal 0.9890478 0.9890478 Vertebrate 0.9851104 0.9851104 Canidae 0.9813780 0.9813780 Dog breed 0.9683250 0.9683250 Puppy 0.9400384 0.9400384Golden retriever 0.8966703 0.8966703
Rocker
rocker-org/rocker provides a series of docker images for various R development purposes.
Bioconductor docker intro
“With Bioconductor containers, we hope to enhance Reproducibility: If you run some code in a container today, you can run it again in the same container (with the same tag) years later and know that nothing in the container has changed. You should always take note of the tag you used if you think you might want to reproduce some work later.”
Rscript
Capture arguments in an Rscript on windows and write them to a file
"C:\Program Files\R\R-3.5.0\bin\Rscript.exe" --verbose -e "args = commandArgs(trailingOnly=TRUE)" -e "writeLines(args,'C:\\Dev\\args.txt')" "file1.csv" "file2.csv" "file3.csv"Arguments can be extracted one by one with args[1] commandArgs() returns a character vector containing the name of the executable and the user-supplied command line arguments.
Set knitr options
Those 2 commands are different. Sets the options for chunk, within a knitr chunk inside the .Rmd document
opts_chunk$set(fig.width=10)Sets the options for knitr outside the .Rmd document
opts_knit$set()Manipulate vectors
Cut and split
Cut a vector in smaller components
cut(0:16, 4)Combine vectors
Combine vectors with c()
x <- c(1, 2, 3)
y <- c(4, 5)
c(x, y)Unite multiple columns into one by pasting strings together
df <- expand_grid(x = c("a", NA), y = c("b", NA))
df
df %>% unite("z", x:y, remove = FALSE)
# To remove missing values:
df %>% unite("z", x:y, na.rm = TRUE, remove = FALSE)
# Separate is almost the complement of unite
df %>%
unite("xy", x:y) %>%
separate(xy, c("x", "y"))
# (but note `x` and `y` contain now "NA" not NA)Maps
Choropleth maps
Shape files to make choropleth maps of Europe: GISCO: Geographical Information and maps
Gisco R related to the above Eurostat package
Other potentially interesting packages:
Bounding boxes
Find the position of countries close to the edge of the map
for (country in c("Portugal", "Finland", "Romania", "Italy")){
print(country)
print(st_bbox(nuts0[nuts0$country == country, ]))
}Map from geojson files
https://r-graph-gallery.com/325-background-map-from-geojson-format-in-r.html
library(sf) library(ggplot2) my_sf <- read_sf(tmp_geojson) my_sf_region_6 <- my_sf[substr(my_sf$code, 1, 2) == “06”, ] ggplot(my_sf_region_6) + geom_sf(fill = “#69b3a2”, color = “white”) + theme_void()
Packages
You might want to read the CRAN manual on Writing R Extensions, and its section on Package dependencies. See also Hadley’s book on R package and its section on Namespace
Use the devtools library to start a package folder structure:
devtools::create("package_name")Use git to track code modifications (shell commands):
$ cd package_name
$ git initDocumentation
The roxygen2 package helps with function documentation. Documentation
can be written in the form of comments #’ tags such as
@param and @description structure the
documentation of each function.
For an introcution to roxygen2, call
vignette("roxygen2", package = "roxygen2") at the R
prompt.
Since roxygen2 version 6, markdown formating can be used in the
documentation, by specifying the @md tag.
Examples
Examples are crucial to demonstrate the use of a fonction. They can
be specified in a roxygen block called @examples:
#' @examplesWrap the examples in donttest if you don’t want R CMD check to test them at package building time.
#' \donttest{dIt is also possible to wrap them in another statement called
dontrun, but this is not recomended on CRAN according to
this Stackoverflow
question.
Vignettes
Vignettes: long-form documentation
devtools::use_vignette("my-vignette")Where to put package vignettes for CRAN submission
“You put the .Rnw sources in vignettes/ as you did, but you missed out a critical step; don’t check the source tree. The expected workflow is to build the source tarball and then check that tarball. Building the tarball will create the vignette PDF.”
R CMD build ../foo/pkg
R CMD check ./pkg-0.4.tar.gzIssues while building vignette for a packages:
sh: 1: /usr/bin/texi2dvi: not found
sudo apt-get install texinfo
“Maybe you’re running R CMD check using the directory name rather than the .tar.gz file?”
“Installing texlive-fonts-extra should take care of it.”
R CMD checking data for non-ASCII characters found 179 marked UTF-8 strings No solution for this one but I guess it’s ok since it concerns country names?
Unit tests
Back in R, add testing infrastructure:
devtools::use_testthat()When checking the package with R CMD CHECK, How can I handle R CMD check “no visible binding for global variable”? These notes are caused by variables used with dplyr verbs and ggplot2 aesthetics.
Continuous Integration
It is good to know if your package can be installed on a fresh system. Continuous integration systems make this possible each time you submit a modification to your repository.
I have used travis-ci which is free for open github repositories. Instructions to
build an R project on travis. Unit tests are also run on travis, in
addition to R CMD CHECK. Package dependencies can be configured in a
.travis.yml file that is read by the travis machine
performing the build. For package that are not on Cran, it’s possible to
specify a dependency field under r_github_packages.
In 2020 RopenSci moved away from Travis CI because of a change of mannagement which removed support for open source projects. In this post Jeroen Ooms thanks Travis CI for greatly enabling CI since 2012.
Make file
- kbroman minimal make
- Cran how to use make files with R CMD build
I copied the make file from the knitr package.
# prepare the package for release
PKGNAME := $(shell sed -n "s/Package: *\([^ ]*\)/\1/p" DESCRIPTION)
PKGVERS := $(shell sed -n "s/Version: *\([^ ]*\)/\1/p" DESCRIPTION)
PKGSRC := $(shell basename `pwd`)
all: docs check clean install
deps:
tlmgr install pgf preview xcolor;\
Rscript -e 'if (!require("Rd2roxygen")) install.packages("Rd2roxygen", repos="http://cran.rstudio.com")'
docs:
R -q -e "devtools::document(roclets = c('rd', 'collate', 'namespace', 'vignette'))"
build:
cd ..;\
R CMD build $(PKGSRC)
install: build
cd ..;\
R CMD INSTALL $(PKGNAME)_$(PKGVERS).tar.gz
check: build
cd ..;\
R CMD check $(PKGNAME)_$(PKGVERS).tar.gz --as-cran
clean:
cd ..;\
$(RM) -r $(PKGNAME).Rcheck/Submitting to CRAN
Before submitting to CRAN.
Update the date and version number in the DESCRIPTION file
Build the package with the make file. The make file will also run the same checks that CRAN will run, so make sure to fix any issues reported there. The following command runs checks on the package’s
.tar.gzarchive. It is one of the commands inside the makefile:R CMD check \((PKGNAME)_\)(PKGVERS).tar.gz –as-cran
Submit package to CRAN The second page will ask to
Step 1 (Upload) the
*.tar.gzfile.Step 2 (Submission) review information from the package’s DESCRIPTION file.
Step 3 (Confirmation)
The maintainer of this package has been sent an email to confirm the submission. After their confirmation the package will be passed to CRAN for review.
In general enter the Git tag after the submission has been completed, because it is very likely that additional information will need to be added to the package before or during the submission process.
CRAN’s automated check of URL’s validity
Upon submission CRAN runs an automated check of URL validity. This blog post explains that URLS in vignettes and function documentations are checked.
Is it possible to rename a package on CRAN
Uwe Ligges on the R-devel mailing list:
“Renaming packages should be reduced to an absolute minimum and would probably only accepted for legal concerns related to inappropriate package names.”
Plots
- R graph gallery https://r-graph-gallery.com/
Plotting with ggplot2
geom_bar
geom_tile + a gradient produce heat mapsAB lines
Plot a line with a slope of 1, y=x
library(ggplot2)
ggplot() +
geom_abline(slope=1)library(stats)
library(ggplot2)
reg <- lm(mpg ~ wt, data = mtcars)
coeff=coefficients(reg)
# Equation of the line :
eq = paste0("y = ", round(coeff[2],1), "*x + ", round(coeff[1],1))
# Plot
sp <- ggplot(data=mtcars, aes(x=wt, y=mpg)) + geom_point()
sp + geom_abline(intercept = 37, slope = -5)+
ggtitle(eq)Axes, scales
Increase number of axis ticks by specifying an integer break vector
scale_x_continuous(breaks = 1:10)Rotate the tick text
Rotate the tick text of the X axis. SO question rotating axis labels in ggplot2
theme(axis.text.x=element_text(angle = 90, hjust=0))Dates
When dates are on the x axis, use a time scale and specify the breaks as a time vector
scale_x_date(breaks = lubridate::parse_date_time(2000:2020, "%y"))Alternatively breaks can be specified as an interval, and minor breaks as well
scale_x_datetime(date_breaks = "5 years", labels = scales::date_format("%Y"), date_minor_breaks = "1 year")Percentages
To display shares as percentages, use
scales::percent
scale_y_continuous(labels = scales::percent)Legend
Place the legend below a plot:
library(ggplot2)
ggplot(iris, aes(Petal.Width, Sepal.Width, color=Species)) +
geom_point() +
theme(legend.position="bottom")Remove the legend
ggplot(iris, aes(Petal.Width, Sepal.Width, color=Species)) +
geom_point() +
theme(legend.position="none")Remove only part of the legend
guides(colour = "none")Change the name of a particular sub element in the legend:
labs(color="Iris Species")
labs(linetype="Sawnwood product group")
labs(y="million")Make a multi column legend
library(scales)
plot + # With a fill aeasthetic
guides(fill=guide_legend(ncol = 2))
plot + # with a color aesthetic
guides(col = guide_legend(ncol = 1))See ggplot2 reference guide legend.
Change the order of legend items
Reorder items in the fill legend:
library(ggplot2)
ggplot(iris, aes(x=Species, y=Petal.Width, fill=Species)) +
geom_boxplot() +
scale_fill_discrete(breaks= c("virginica", "setosa", "versicolor"))One legend for each sub plot
Different legends for each sub plot using grid extra
library(ggplot2)
library(gridExtra)
df <- data.frame(country = rep(c("X", "Y", "Z"), each=9),
product = rep(c("A", "B", "C", "B", "C", "D", "B", "D", "A"),each=3),
year = rep(1:3,9),
value = rnorm(27)
)
out <- by(data = df,
INDICES = df$product,
FUN = function(m) {
ggplot(m, aes(x=year, y=value, color=country)) +
geom_point() +
facet_wrap(~product, scales="free_y") +
theme_minimal()
})
do.call(grid.arrange, out)
# If you want to supply the parameters to grid.arrange
#do.call(grid.arrange, c(out, ncol=3))Different legend for each sub plot using patchwork
Facet plots
Facet grid
“‘facet_grid()’ forms a matrix of panels defined by row and column faceting variables.”
Facet wrap, specify only the
“‘facet_wrap()’ wraps a 1d sequence of panels into 2d.”
p <- ggplot(mpg, aes(displ, hwy)) + geom_point() # Use vars() to supply faceting variables: p + facet_wrap(vars(class))
Note ?vars:
> ‘vars()’ is superseded because it is only needed for the scoped verbs
> (i.e. ‘mutate_at()’, ‘summarise_at()’, and friends), whichMath formulas in facet labels
Note to add a new line to a facet label, simply insert it with
facet_variable = gsub(" ","\n", facet_variable)library(dplyr)
library(ggplot2)
# Create a first facet variable with examples of math formulas
iris2 <- iris %>%
mutate(species_math = factor(Species,
levels = c("setosa", "versicolor", "virginica"),
labels = c("m^2",
expression(bar(x) == sum(frac(x[i], n), i==1, n) * beta * Q[t-1]),
bquote(pi == .(pi)))))
# Create a second facet variable with mean lengths
# This illustrates how to pass a numeric vector inside a formula
iris_mean <- iris2 %>%
group_by(Species) %>%
summarise(across(ends_with("Length"), mean), .groups="drop")
iris2$mean_length <- factor(iris2$Species,
levels = c("setosa", "versicolor", "virginica"),
labels = mapply(function(p, s) bquote(bar(p) == .(p) ~ bar(s) ==.(s)),
round(iris_mean$Petal.Length,3), round(iris_mean$Sepal.Length,3)))
# Create the faceted plot
iris2 %>%
ggplot(aes(x = Petal.Length, y = Petal.Width)) +
geom_point() +
facet_wrap(species_math ~ mean_length + Species,
labeller = labeller(species_math = label_parsed, mean_length = label_parsed))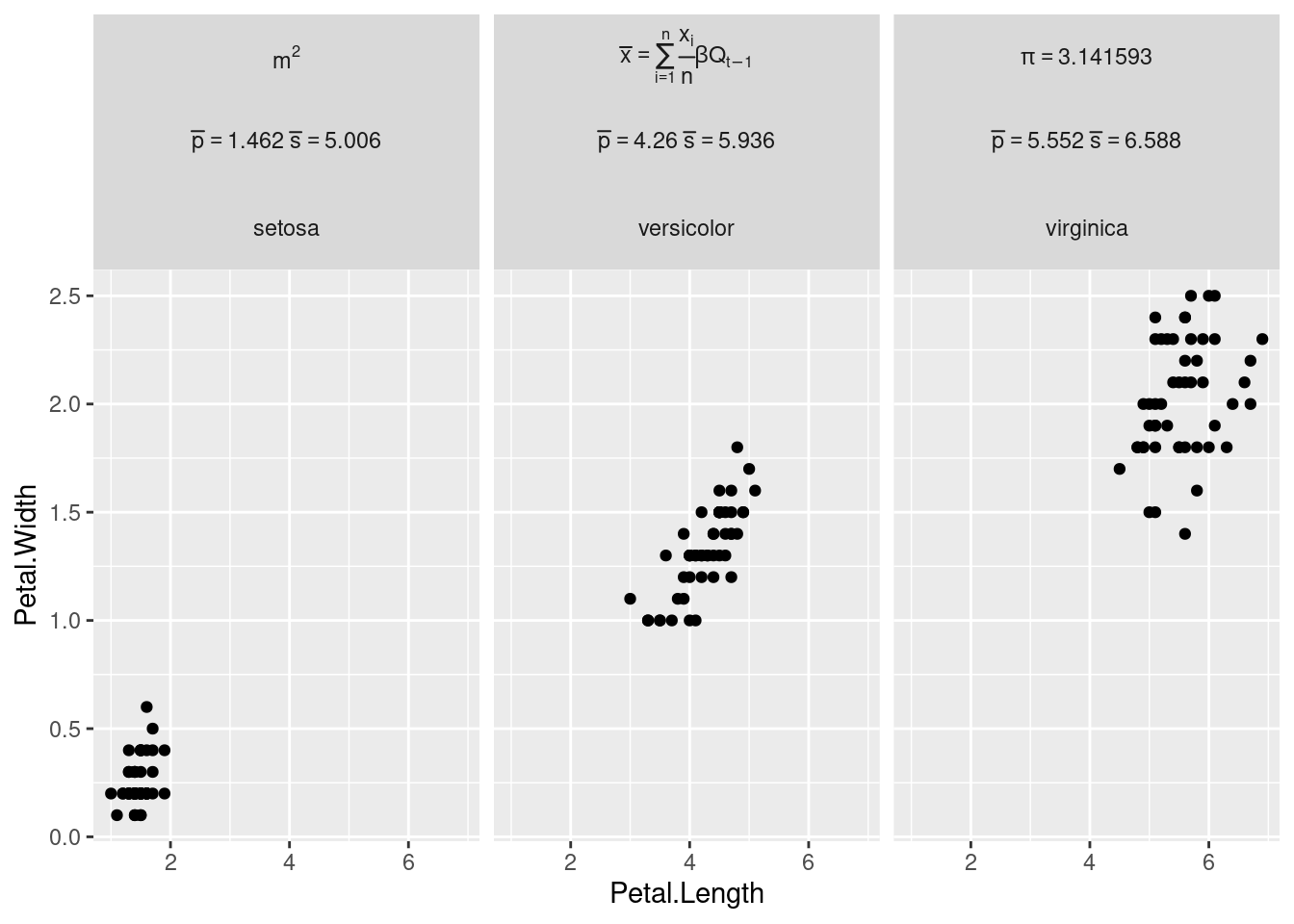
As shown in the example above, the labeller can parse:
A character vector such as “m^2” for simple formulas
An
expressionfor more complex math with indicesThe output of
bquoteto include numerical values in the formula. See also this answer on how to usebquotewith numerical vectors of more than one value.see this other answer on how to apply the labeller to one of the faceting variables only. In our case we apply it to the
species_mathvariable only.
The syntax is different from Latex math formulas because
label_parsed interprets labels as plotmath
expressions. For example indices are written x_i in Latex
and x[i] in plot math expressions, and Greek letters are
written directly as alpha instead of \alpha in
Latex. You can find many formulas in the help page of the plotmath
function. Good luck with the plotmath examples.
Palettes and colours
scale_gradient2 creates a diverging colour gradient (low-mid-high), scale_gradientn creates a n-colour gradient. For binned variants of these scales, see the color steps scales.
For example, I used this fora choropleth map of carbon sink
scale_fill_gradient2(low="#018606", mid="white", high="red")+Themes
Minimal theme
theme_minimal()Geoms
Bar plots with geom_col
Use geom_col for bar plots where the height of the column represent the value of a variable. Use geom_bar for plots where the height of the column represents a count of number of occurrences.
geom_ribbon
Example from help(geom_ribbon):
huron <- data.frame(year = 1875:1972, level = as.vector(LakeHuron))
ggplot(huron, aes(year)) +
geom_ribbon(aes(ymin = level - 1, ymax = level + 1), fill = "grey70") +
geom_line(aes(y = level))Add or remove lines at the border of the ribbon:
ggplot(huron, aes(year)) +
geom_ribbon(aes(ymin = level - 1, ymax = level + 1),
outline.type="lower", fill = "grey70", color="red") +
geom_line(aes(y = level))
ggplot(huron, aes(year)) +
geom_ribbon(aes(ymin = level - 1, ymax = level + 1),
outline.type="lower", fill = "grey70", color="red",
linetype=0) +
geom_line(aes(y = level))Palettes
Setting up colour palettes in R
To create a RColorBrewer palette, use the brewer.pal function. It takes two arguments: n, the number of colors in the palette; and name, the name of the palette. Let’s make a palette of 8 colors from the qualitative palette, “Set2”.
library(RColorBrewer)
brewer.pal(n = 8, name = "Set2")
#[1] "#66C2A5" "#FC8D62" "#8DA0CB" "#E78AC3" "#A6D854" "#FFD92F" "#E5C494" "#B3B3B3"
palette(brewer.pal(n = 8, name = "Set2"))Use this palette in ggplot2
ggplot(iris, aes(x=Sepal.Length, y=Petal.Length, color=Species)) +
geom_point() +
scale_color_brewer(palette = "Set2")Manual colours
Use a named vector to set a palette in ggplot2 as explained in ggplot2 scale_manual
p <- ggplot(iris, aes(x=Sepal.Length, y=Petal.Length, color=Species)) +
geom_point()
p + scale_colour_manual(values = c(setosa='black', versicolor='red', virginica='green'))Create a named palette using R colour brewer:
species_names <- c("setosa", "versicolor", "virginica")
iris_palette <- setNames(brewer.pal(n=length(species_names), name='Set2'),
species_names)
p + scale_colour_manual(values = iris_palette)Display palettes
Display qualitative palettes:
display.brewer.all(type="qual") Display all palettes
display.brewer.all()Add more colours to a palette
Expand from 8 colours to more colours:
mycolors <- colorRampPalette(brewer.pal(8, "Set2"))(nb.cols)Text labels
ggplot2 text
Use position jitter
# Reduce the number of points so it becomes more readable
iris2 <- iris %>%
group_by(Species, Petal.Width) %>%
summarise(Sepal.Width = mean(Sepal.Width))
ggplot(iris2, aes(x = Petal.Width, y=Sepal.Width, color = Species)) +
geom_text(aes(label = Species),
position=position_jitter(width=0.5,height=0.3),
hjust="left") +
# Expand the scale so that names are shown in full
scale_x_continuous(expand = expansion(mult=c(0.1,0.5)))Use repulsive textual annotations
ggplot(iris2, aes(x = Petal.Width, y=Sepal.Width, color = Species)) +
ggrepel::geom_text_repel(aes(label = Species))Change the text size
ggplot(iris2, aes(x = Petal.Width, y=Sepal.Width, color = Species)) +
ggrepel::geom_text_repel(aes(label = Species), size=8)Justification
df <- data.frame(
x = c(1, 1, 2, 2, 1.5),
y = c(1, 2, 1, 2, 1.5),
text = c("bottom-left", "bottom-right", "top-left", "top-right", "center")
)
ggplot(df, aes(x, y)) +
geom_text(aes(label = text))
ggplot(df, aes(x, y)) +
geom_text(aes(label = text), vjust = "inward", hjust = "inward")Python compared to R
See also the python page for a comparison between pandas methods and R data frames functions.
l = c(1,2,3)
s = l
s[3]
[1] 3
s[3] = "a"
s
[1] "1" "2" "a"
l
[1] 1 2 3Using the address function to see the address of these objects in memory We can see that s and l share the same address. The address only changes when we asign something to s.
library(pryr)
l = c(1,2,3)
s = l
address(l)
[1] "0x316f718"
address(s)
[1] "0x316f718"
s[3] = "a"
address(s)
[1] "0x36a7d30"
s
[1] "1" "2" "a"
l
[1] 1 2 3Checking string objects
bla = "qsdfmlkj"
address(bla)
[1] "0x38e5120"
bli = bla
address(bli)
[1] "0x38e5120"
bli = paste(bli, "sdf")
address(bli)
[1] "0x38d1d60"TODO Compare to the same code in python to see the difference between the above and passing by reference.
Shiny
App.R
“Note: Prior to version 0.10.2, Shiny did not support single-file apps and the ui object and server function needed to be contained in separate scripts called ui.R and server.R, respectively. This functionality is still supported in Shiny, however the tutorial and much of the supporting documentation focus on single-file apps.”
Tidyverse
dplyr
Group_by and summarise
library(dplyr)
cars %>%
group_by(speed) %>%
print(n=2) %>% # works because the print function returns its argument
summarise(numberofcars = n(),
min = min(dist),
mean = mean(dist),
max = max(dist)) ## # A tibble: 50 × 2
## # Groups: speed [19]
## speed dist
## <dbl> <dbl>
## 1 4 2
## 2 4 10
## # ℹ 48 more rows## # A tibble: 19 × 5
## speed numberofcars min mean max
## <dbl> <int> <dbl> <dbl> <dbl>
## 1 4 2 2 6 10
## 2 7 2 4 13 22
## 3 8 1 16 16 16
## 4 9 1 10 10 10
## 5 10 3 18 26 34
## 6 11 2 17 22.5 28
## 7 12 4 14 21.5 28
## 8 13 4 26 35 46
## 9 14 4 26 50.5 80
## 10 15 3 20 33.3 54
## 11 16 2 32 36 40
## 12 17 3 32 40.7 50
## 13 18 4 42 64.5 84
## 14 19 3 36 50 68
## 15 20 5 32 50.4 64
## 16 22 1 66 66 66
## 17 23 1 54 54 54
## 18 24 4 70 93.8 120
## 19 25 1 85 85 85group_by() creates a tbl_df objects which is a wrapper around a data.frame to enable some functionalities. Note that print returns its output on a tbl_df object. So print() can be used inside the pipe without stopping the workflow.
Count and tally
Count the unique values of one or more variables
count(iris, Species, sort=TRUE)is equivalent to
iris %>%
group_by(Species) %>%
tally(sort=TRUE)The wt argument can be used to perform weighted
counts
count(iris, Species, wt=Sepal.Length, sort=TRUE)Sum across multiple columns
https://stackoverflow.com/questions/29006056/efficiently-sum-across-multiple-columns-in-r
Across
Apply the same transformation to multiple columns, allowing to use select semantics inside summarise and mutate.
cars %>%
group_by(speed) %>%
summarise(across(dist, list(n = length , min = min, mean = mean, max = max)))## # A tibble: 19 × 5
## speed dist_n dist_min dist_mean dist_max
## <dbl> <int> <dbl> <dbl> <dbl>
## 1 4 2 2 6 10
## 2 7 2 4 13 22
## 3 8 1 16 16 16
## 4 9 1 10 10 10
## 5 10 3 18 26 34
## 6 11 2 17 22.5 28
## 7 12 4 14 21.5 28
## 8 13 4 26 35 46
## 9 14 4 26 50.5 80
## 10 15 3 20 33.3 54
## 11 16 2 32 36 40
## 12 17 3 32 40.7 50
## 13 18 4 42 64.5 84
## 14 19 3 36 50 68
## 15 20 5 32 50.4 64
## 16 22 1 66 66 66
## 17 23 1 54 54 54
## 18 24 4 70 93.8 120
## 19 25 1 85 85 85With trade data
summarise(across(all_of(c("tradevalue", "weight", "quantity")), sum, na.rm=TRUE))Other examples from the help of the across function
# A purrr-style formula
iris %>%
group_by(Species) %>%
summarise(across(starts_with("Sepal"), ~mean(.x, na.rm = TRUE)))## # A tibble: 3 × 3
## Species Sepal.Length Sepal.Width
## <fct> <dbl> <dbl>
## 1 setosa 5.01 3.43
## 2 versicolor 5.94 2.77
## 3 virginica 6.59 2.97# A named list of functions
iris %>%
group_by(Species) %>% #
summarise(across(starts_with("Sepal"), list(mean = mean, sd = sd)))## # A tibble: 3 × 5
## Species Sepal.Length_mean Sepal.Length_sd Sepal.Width_mean Sepal.Width_sd
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 setosa 5.01 0.352 3.43 0.379
## 2 versicolor 5.94 0.516 2.77 0.314
## 3 virginica 6.59 0.636 2.97 0.322# Show all_of to select a vector of variables
iris %>%
group_by(Species) %>%
summarise(across(all_of(c("Sepal.Length", "Petal.Width")),
~mean(.x, na.rm = TRUE)))## # A tibble: 3 × 3
## Species Sepal.Length Petal.Width
## <fct> <dbl> <dbl>
## 1 setosa 5.01 0.246
## 2 versicolor 5.94 1.33
## 3 virginica 6.59 2.03More information in the vignette
vignette("colwise", package="dplyr")The vignette mentions this about rename:
“across() doesn’t work with select() or rename() because they already use tidy select syntax; if you want to transform column names with a function, you can use rename_with().”
Filter
Filter a single species or two species
library(dplyr)
iris %>% filter(Species == "setosa")
iris %>% filter(Species %in% c("setosa", "virginica"))Filter rows where the variable contain 2 characters only
nuts0 <- nuts %>% filter(nchar(nuts$nuts_id) == 2)Mutate to create a new vector
Mutate multiple variables in the dataframe at once using the
vars() helper function to scope the mutation: Note
mutate_at has been superseded by the across function.
iris %>%
mutate_at(vars(starts_with("Petal")), round) %>%
head()## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1 0 setosa
## 2 4.9 3.0 1 0 setosa
## 3 4.7 3.2 1 0 setosa
## 4 4.6 3.1 2 0 setosa
## 5 5.0 3.6 1 0 setosa
## 6 5.4 3.9 2 0 setosaRename columns
rename_with from the dplyr package can use
either a function or a formula to rename a selection of columns given as
the .cols argument. For example passing the function name
toupper or tolower:
library(dplyr)
rename_with(head(iris), toupper, starts_with("Petal"))
rename_with(head(iris), tolower, starts_with("Petal"))Is equivalent to passing the formula ~ toupper(.x):
rename_with(head(iris), ~ toupper(.x), starts_with("Petal"))To rename all columns, you can also use set_names from
the rlang package. We now use paste0 as a renaming
function. pasteO takes 2 arguments, as a result there are
different ways to pass the second argument depending on whether we use a
function or a formula.
rlang::set_names(head(iris), paste0, "_hi")
rlang::set_names(head(iris), ~ paste0(.x, "_hi"))The same can be achieved with rename_with by passing the
data frame as first argument .data, the function as second
argument .fn, all columns as third argument
.cols=everything() and the function parameters as the
fourth argument .... Alternatively you can place the
second, third and fourth arguments in a formula given as the second
argument.
rename_with(head(iris), paste0, everything(), "_hi")
rename_with(head(iris), ~ paste0(.x, "_hi"))rename_with only works with data frames.
set_names is more generic and can also perform vector
renaming
rlang::set_names(1:4, c("a", "b", "c", "d"))Recode values in a vector
“Replace numeric values based on their position or their name, and character or factor values only by their name.”
char_vec <- sample(c("a", "b", "c"), 10, replace = TRUE)
recode(char_vec, a = "Apple")Pipes
purrr
Hadley Wickham’s answer to a SO question Why use purrr::map instead of lapply?
Map a function to nested data sets
Load data
list.files(getwd())
forestEU_wide <- read.csv("Forest-R-EU.csv", stringsAsFactors = FALSE)
head(forestEU)Pivot to long format
# at some pointin the future this will be called pivot_long
forestEU <- forestEU_wide %>%
# select everything except the Year, then pivot all columns and put the value in area
gather(-Year, key = "Country", value = "Area")Tricks
David Robinson Ten tricks in the tidyverse
- count with weight, sort and renamining count(x, …, wt = NULL, sort = FALSE, name = “n”)
- creating variables in count()
- add_count()
- summarize with a list column (to create many models for example)
- fct_reorder() + geom_col() + coord_flip()
- fct_lump() to lump less frequent values together
- use a log scale scale_x/y_log10
- crossing()
- separate()
- extract() use a regular expression such as
S(.*)E(.*)to extract series and episode from a string of the form “S32E44”.
Purrr
Interpolation
Interpolate for one country
country_interpolation <- function(df) {
df <- data.frame(approx(df$Year, df$Area, method = "linear", n=71))
df <- rename(df, Year = x, Area = y)
return(df)
}
forestEU %>%
filter(Country=="Austria") %>%
country_interpolation()Interpolate for all countries Perform the Interpolation on all countries
See documentation in * many models https://r4ds.had.co.nz/many-models.html * blog https://emoriebeck.github.io/R-tutorials/purrr/
forestEU_nested <- forestEU %>%
# Remove empty area
filter(!is.na(Area)) %>%
group_by(Country) %>%
nest() %>%
mutate(interpolated = map(data, country_interpolation))
# forestEU_nested %>% unnest(data)
# Unnest the interpolated data to look at it and plotting
forestEU_interpolated <- forestEU_nested %>% unnest(interpolated)Read many csv
See iris_csv file created in the csv section of this document. Read these csv files into one data frame. Note the Species column has to be present in the csv files, otherwise we would loose that information.
iris_csv <- list.files(data_folder, full.names = TRUE) %>%
purrr::map_dfr(read_csv)Write to many csv
#dir.create("output")
forestEU_nested <- forestEU_nested %>%
mutate(filename = paste0("output/", Country, ".csv"),
wrote_stuff = map2(interpolated, filename, write.csv))tidy evaluation
scatter_plot <- function(data, x, y) {
x <- enquo(x)
y <- enquo(y)
ggplot(data) + geom_point(aes(!!x, !!y))
}
scatter_plot(mtcars, disp, drat)Another example use of metaprogramming to change variables
add1000 <- function(dtf, var){
varright <- enquo(var)
varleft <- quo_name(enquo(var))
dtf %>%
mutate(!!varleft := 1000 + (!!varright))
}
add1000(iris, Sepal.Length)Tidy select
Tidy select selection
language explains the various selection function such as
all_of, contains and
starts_with.
library(dplyr)
iris2 <- head(iris, 1)
select(iris2, contains("w")) # Literal string
select(iris2, matches("w")) # Regular expression
select(iris2, starts_with("S"))
select(iris2, all_of(c("Sepal.Length", "Petal.Width")))The selection can be inverted by using s minus sign in front of it. Not it’s not a question mark as explained in this SO answer because the selection fonctions return numerical vectors of integer column positions (not boolean values).
library(dplyr)
select(head(iris), -matches("w") )tidyr
tidyr vignette on tidy data In the section on “Multiple types in one table”:
Datasets often involve values collected at multiple levels, on different types of observational units. During tidying, each type of observational unit should be stored in its own table. This is closely related to the idea of database normalisation, where each fact is expressed in only one place. It’s important because otherwise inconsistencies can arise.
Normalisation is useful for tidying and eliminating inconsistencies. However, there are few data analysis tools that work directly with relational data, so analysis usually also requires denormalisation or the merging the datasets back into one table.
Example use of tidyr::nest() to generate a group of
plots: make
ggplot2 purrr.
library(tidyr)
library(dplyr)
library(purrr)
library(ggplot2)
piris <- iris %>%
group_by(Species) %>%
nest() %>%
mutate(plot = map2(data, Species,
~ggplot(data = .x,
aes(x = Petal.Length, y = Petal.Width)) +
geom_point() + ggtitle(.y)))
piris$plot[1]
piris$plot[3]
piris$plot[2]Time series
Moving averages
Use the rollmean function from the zoo package. It defaults to centered.
library(zoo)
x.Date <- as.Date(paste(2004, rep(1:4, 4:1), sample(1:28, 10), sep = "-"))
x <- zoo(rnorm(12), x.Date)
rollmean(x, 3, fill=NA)
rollmean(x, 3, fill="extend")
y <- x
y["2004-01-15"] <- NA
rollmean(y, 3, fill="extend")
# Returns NA values if there are not enough observations for the rollmean
rollmean(y, 9, fill="extend")
rollmean(x, 9, fill="extend") Working directory
Hadley Wickham on a Stackoverflow comment:
“You should never use setwd() in R code - it basically defeats the idea of using a working directory because you can no longer easily move your code between computers. – hadley Nov 20 ’10 at 23:44”
Content copied from my StackOverflow answer.
TLDR: The here
package (available on CRAN) helps you build a path from a project’s root
directory. R projects configured with here() can be shared
with colleagues working on different laptops or servers and paths built
relative to the project’s root directory will still work. The
development version is at github.com/r-lib/here.
With git
You certainly store your R code in a directory. This directory is probably part of a git repository and/or an R studio project. I would recommend building all paths relative to that project’s root directory. For example let’s say that you have an R script that creates reusable plotting functions and that you have an R markdown notebook that loads that script and plots graphs in a nice (so nice) document. The project tree would look something like this
├── notebooks
│ ├── analysis.Rmd
├── R
│ ├── prepare_data.R
│ ├── prepare_figures.RFrom the analysis.Rmd notebook, you would import
plotting function with here() as such:
source(file.path(here::here("R"), "prepare_figures.R"))Why?
From the Ode to the here package:
Do you: Have setwd() in your scripts? PLEASE STOP DOING THAT. This makes your script very fragile, hard-wired to exactly one time and place. As soon as you rename or move directories, it breaks. Or maybe you get a new computer? Or maybe someone else needs to run your code? […] Classic problem presentation: Awkwardness around building paths and/or setting working directory in projects with subdirectories. Especially if you use R Markdown and knitr, which trips up alot of people with its default behavior of “working directory = directory where this file lives”. […]
Install the here package:
install.packages("here")
library(here)
here()
here("construct","a","path")Documentation of the here() function:
Starting with the current working directory during package load time, here will walk the directory hierarchy upwards until it finds a directory that satisfies at least one of the following conditions:
- contains a file matching [.]Rproj$ with contents matching ^Version: in the first line
- [… other options …]
- contains a directory .git
Once established, the root directory doesn’t change during the active R session. here() then appends the arguments to the root directory.
The development version of the here package is available on github.
What about
What about files outside the project directory?
If you are loading or sourcing files outside the project directory, the recommended way is to use an environment variable at the Operating System level. Other users of your R code on different laptops or servers would need to set the same environment variable. The advantage is that it is portable.
data_path <- Sys.getenv("PROJECT_DATA")
df <- read.csv(file.path(data_path, "file_name.csv"))Note: There is a long list of environmental variables which can affect an R session.
What about many projects sourcing each other?
It’s time to create an R package.
Further resources
Blogs
tinyverse “Lightweight is the right weight”
Dirk Eddelbuettel dependencies
“I relied on the liteq package by Gabor which does the job: enqueue jobs, and reliably dequeue them (also in a parallel fashion) and more. It looks light enough:”
R> tools::package_dependencies(package="liteq", recursive=FALSE, db=AP)$liteq [1] "assertthat" "DBI" "rappdirs" "RSQLite"“Two dependencies because it uses an internal SQLite database, one for internal tooling and one for configuration.” All good then? Not so fast. The devil here is the very innocuous and versatile RSQLite package because when we look at fully recursive dependencies all hell breaks loose: Now we went from four to twenty-four, due to the twenty-two dependencies pulled in by RSQLite.”
Gavin Simpson My aversion to pipers shows an Hadley tweet explaining that pipe might not be good in package development.
5 R packages for text analysis https://towardsdatascience.com/r-packages-for-text-analysis-ad8d86684adb
Shiny apps by international organizations
- The human mortality database short term mortality fluctuations visualization toolkit, Git repository, Plos One paper
Stackoverflow
- Stack overflow question checklist general programming
- How to make a great R reproducible example